The profiling of individual cells within a heterogeneous cellular population is vital to advancing research across numerous biological and biomedical fields. Unlike bulk methods, single-cell techniques can capture the full diversity of the cellular landscape and allow for the investigation of distinct gene expression patterns. Current methods of measuring RNA expression in single cells, such as by using fluorescence-activated cell sorting (FACS) to isolate cells in 96-well microtiter plates followed by reverse transcription polymerase chain reaction (RT-PCR), have not been widely adopted due to low throughput and high associated costs, or, in the case of flow-cytometry based fluorescence in situ hybridization (FISH-flow), due to time-consuming sample preparation. Using valve-operated microfluidics to isolate single cells for RT-PCR offers the advantage of a higher throughput, but still requires a sophisticated microfluidics setup that may be expensive, difficult to operate, and prone to errors. A droplet microfluidics approach would allow for ultra-high-throughput single cell RT-PCR (scRT-PCR), but ideally would need to be able to isolate and retain encapsulated cells and biomolecules throughout multi-step assays using standard laboratory equipment. Here, the authors present a high-throughput scRT-PCR technique that profiles single cells isolated in semi-permeable microcapsules. Once the semi-permeable capsules (SPCs) have been formed, multi-step analytical procedures can be carried out with only a regular pipette and tubes, and the SPCs are compatible with conventional FACS for single-cell RNA cytometry.
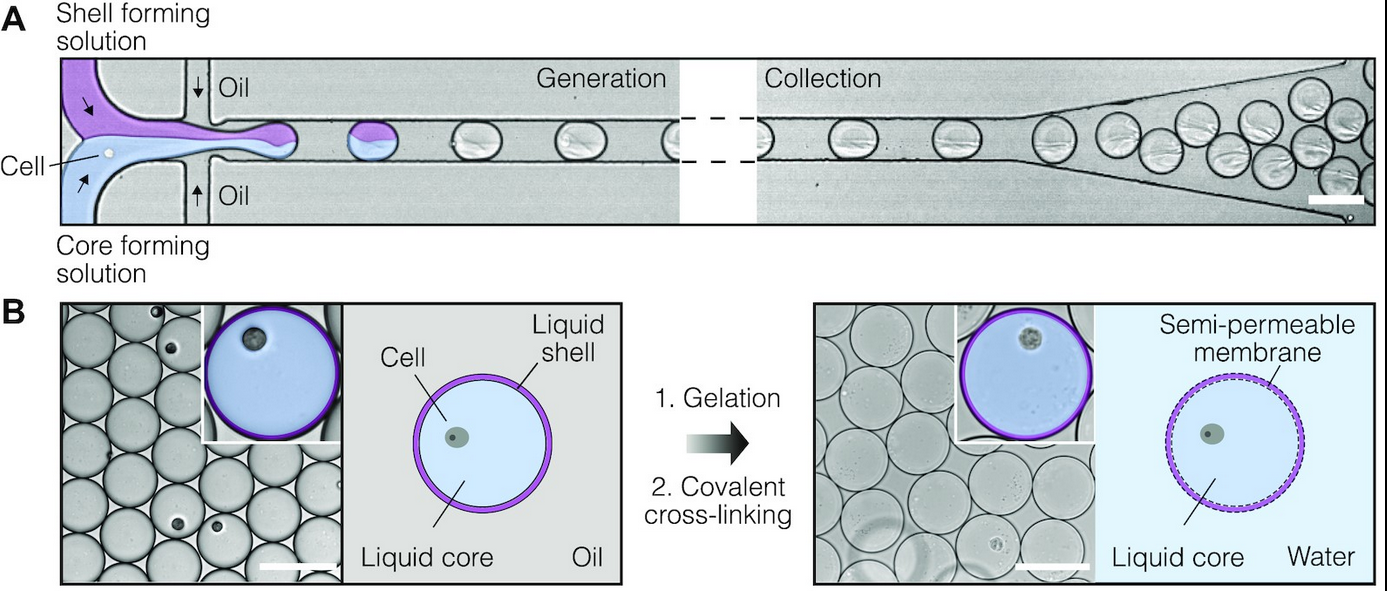
Figure 2 A&B
The SPCs are composed of aqueous two-phase system droplets in oil. Cells are suspended in a dextran solution at an appropriate density and infused with gelatin methacrylate during droplet generation. Upon liquid-liquid phase separation, the dextran solution forms a liquid core containing no more than one cell, and the gelatin methacrylate forms a liquid shell around this core. The liquid shell is then cooled to a gel and covalently cross-linked by photopolymerization, resulting in a stable, semi-permeable membrane. By encapsulating double-stranded DNA fragments of assorted sizes and examining DNA retention after the SPCs were washed, it was determined that fragments ≥300 bp (MW ~180 kDa) were retained within this membrane. This permeability cutoff is well-suited for many biological applications, as it would allow for the exchange of reagents in multi-step assays and the removal of intracellular proteins (e.g., RNases) while retaining larger biomolecules, whole cells, and viruses. The authors also note that the microcapsule’s membrane structure can be fine-tuned for specific applications that require different permeability cut-offs. In addition to their permeability, SPCs remain intact during freezing, thawing, centrifugation, and exposure to various salts and solvents (e.g., 4M guanidinium isothiocyanate, 90% methanol, 90% acetone). Finally, in contrast with hydrogel bead-based assays, there was no significant cell loss detected during the microcapsule generation process, ensuring that the compartmentalized cells are retained for subsequent analysis.
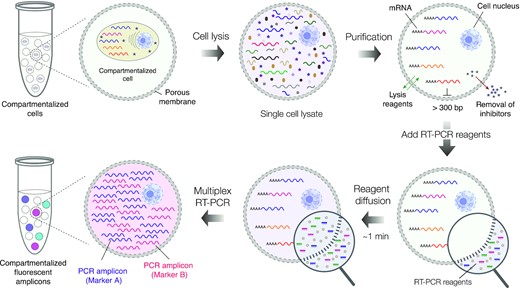
Figure 1
To assess the use of SPCs for profiling single cells based on their gene expression, the authors encapsulated single cells from the human leukemia cell line K-562, single cells from the human embryonic kidney cell line HEK293, and single cells from a 1:1 mixture of K-562 and HEK293 cells. After cell lysis, genomic DNA digestion, and several washes, the microcapsules contained purified total RNA from single cells. The SPCs were then dispersed in an RT-PCR reaction mix that contained fluorescently labelled oligonucleotides targeting transcripts of the protein tyrosine phosphatase receptor type C (PTPRC; specific to K-562 cells), Yes-associated protein 1 (YAP; specific to HEK293 cells), and the cDNA of β-actin (ACTB; ubiquitous expression). Fluorescence microscopy was used to measure the fluorescence intensity of the SPCs; microcapsules containing single K-562 cells (PTPRC+ACTB signals) appeared cyan, microcapsules containing single HEK293 cells (YAP + ACTB signals) appeared magenta, and microcapsules lacking a nucleic acid template remained blank. This allowed for digital gene expression profiling of single cells, and image analysis enabled accurate cell-type identification based on these gene expression profiles. Surprisingly, however, the presence of a third group of microcapsules positive for only the ACTB signal (appeared blue) was noticed. The substantial number of these events compared to the number of loaded cells suggested that these results may be false positive events resulting from ambient RNA molecules that were present in the initial cell suspension. As accurately distinguishing true positive and false positive events is essential, the authors repeated the experiment using mild-lysis conditions aimed at keeping the cell nucleus intact. Indeed, under these conditions the cell nucleus exhibited localized fluorescence within microcapsules whereas microcapsules with ambient RNA displayed diffuse fluorescence, allowing true and false positive results to be clearly differentiated.
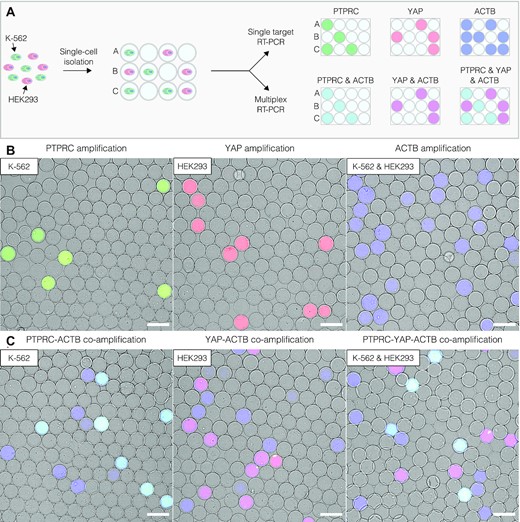
Figure 3
Finally, to validate these microscopy results and to assess the compatibility of the SPC approach with high-throughput flow cytometry instruments, the post-RT-PCR microcapsules were loaded onto the FACS instrument for RNA cytometry. Comparative analysis showed a strong concordance between the microscopy and flow cytometry results, with PTPRC positive events detected in 3.19% and 3.63% of microcapsules and YAP-positive events detected in 4.84% and 5.46% of microcapsules, approaching the theoretical expected values from a Poisson distribution (~5-6%). Co-encapsulation of K-562 and HEK293 was very rare (0.02-0.15%) as expected by the Poisson distribution. Single-cell digital profiles based on PTPRC and YAP gene activity were created and demonstrated the compatibility of the SPC approach with FACS for the ultra-high-throughput gene expression screening of single cells. To demonstrate one potential biomedical application of this approach, primary human peripheral blood mononuclear cells (PMBCs) were mixed with cells from the acute promyelocytic leukemia line NB-4, encapsulated, and dispersed in an RT-PCR mix that contained fluorescently labelled oligonucleotides targeting transcripts of PTPRC and PML-RARα. Leukemic cells could in this way be identified among the mixture of PMBCs, although the choice of targeted transcript may affect the detection rate of these cells. Altogether, the SPC-based RNA cytometry approach described here was fast, simple, and highly sensitive at producing gene expression profiles of thousands of individual cells. The semi-permeable microcapsules offer numerous benefits over the current approaches to single-cell research and demonstrate potential for a broad range of clinical and biological applications in the future.


